Intro to Python
🔎Workshop at a Glance:
The overall goal of the is to learn how to program in Python using modern, reproducible tools.
Session 1 will help you set up a flexible, interactive, working environment for Python programming (Miniconda, VS Code IDE, Jupyter Notebook, etc.)
Session 2 and 3 will focus on the Python basics such as data structure, list comprehensive, and functions. We will also learn data frame manipulations with
pandas.Session 4 will introduce you to object-oriented programming (OOP) with examples from the machine learning library
scikit-learn.
❗What You’ll Need for the Workshop
Bring your laptop (Windows/MacOS)
Basic GitHub knowledge & MSK GitHub Enterprise account–The workshop website is hosted on MSK Enterprise GitHub, which might require you logging in with MSK credentials to access links/download files. Check out Biostatistics Resource Guide for GitHub-related training.
Install Miniconda and Visual Studio Code before session 1 (see this guide)
ImportantWhen following the installation instructions in this article, we recommend that you install software on your MSK laptop or workstation. Downloading/installing software files on VDI is extremely slow and might not install at all if the software is too large.
📖 About this Guide
This handout is a reading for the first session of the Introduction to Python workshop. It serves as a follow-along guide to help you install Python, set up essential tools like Miniconda and VS Code, and prepare for coding in Python for the upcoming sessions.
Along the way, we will also discuss some important questions–What makes Python useful? Why would we, as biostatisticians, want to learn it? You will get an overview of the key features of Python, what it can do in relation to biostatistics/bioinformatics research, and how it compared to R. In the upcoming sessions, we will dive deeper into some of Python’s features and functions through hands-on programming practices.
1. Introduction to Python
What is Python?

Python is a high-level, interpreted, and general-purpose programming language first developed by Guido van Rossum in 1991.
Python has gained much popularity in the past 20 years. Its user group has expanded into a large and active scientific computing and developer community that spans numerous academic and industrial fields. Nowadays, Python has a powerful ecosystem of external packages (libraries) for data science, artificial intelligence, and software development.
Python is cross-platform and open-source. In Python, you can easily install packages with the built-in installer pip or package manager conda, just as you do with install.packages() in R.
What can Python do?
Just like R, Python is an open source and versatile programming language that allows users to perform a wide range of data analysis and computational tasks. While R is particularly useful in statistical analysis and visualizations, Python has been used in many distinct areas, such as:
Machine Learning and Deep learning
Web Development
Scripting & Automation
Cloud Computing
Game Development
Cybersecurity
Why learn Python–as Biostatisticians?
Within the field of biostatistics/ bioinformatics, Python has become a core tool for biomedical data analysis due to its versatility, reproducibility, and strong ecosystem of scientific libraries. Below are some areas where Python can be useful and some essential libraries.
Statistical analysis
While R is the go-to tool for statistical analysis, Python has caught up with many equivalent libraries and functions:
statsmodels/scipy.statsprovide regression modeling and hypothesis testing.lifelines/scikit-survivalsupport survival analysis and plotting.
ML/DL ecosystem
Python dominates in machine learning and AI development:
scikit-learnis a rich machine learning library that supports both supervised regression an dclassification (e.g., random forests, gradient boosting) and unsupervised clustering (e.g., K-means).TensorFlow,PyTorchare deep learning libraries widely used for computer vision and natural language processing.optuna,Raycan be integrated into ML/DL workflows for easy and efficient model training, hyperparameter tuning, fine-tuning, etc.
Omics data analysis
Emerging packages that provide standard omics data preprocessing and analysis pipelines allow Python to become increasingly popular in the field of bioinformatics:
scanpy,anndataare libraries for single-cell RNA-seq data loading, preprocessing, and analysis.Biopythonis a set of tools for biological computation that performs file parsering (BLAST, FASTA, GenBank, etc.), sequence analysis, clustering algorithms, etc.pysamworks with BAM/SAM/VCF files.
Python vs. R: Differences
While both programming languages are popular for data analysis and computation, Python and R differ in their underlying code structure, the scope of functionality, and the extensibility of tasks they can perform. Here is a non-exhaustive summary of some key differences:
| Feature/Task | R | Python |
|---|---|---|
| Programming logic | Mostly function-oriented | Function and object-oriented – structured around classes |
| General-purpose programming | ⚠️ Less ideal – designed mainly for working with data | ✅ Strong – ML & AI, software development, scripting, etc. |
| Computational power | ✅ Vectorization allows operating on all elements of a vector at once ✅ Best for statistical analysis ⚠️ Memory-intensive; often slow for reading large data and performing large computations |
✅ Generally faster for loops ✅ Strong support for GPU computing ✅ Memory-efficient for handling large objects and complex computations |
| Package availability | ✅ Excellent for statistical analysis (glm, survival, ggplot2) ⚠️ Good options for ML (caret, mlr3) but few DL packages ✅ Great for omics-focused analysis (Bioconductor, ComplexHeatmap, Seurat) |
☑️ Improving on statistical packages (statsmodels, lifelines) ✅ Best for ML/DL (scikit-learn, pytorch, keras) ✅ Established packages specialized in processing large omics datasets (scanpy, scvi-tools) |
| IDE & Reproducible environments (notebooks etc.) | ✅ RStudio ✅ RMarkdown, Quarto |
✅ Visual Studio Code, JupyterLab, PyCharm, Spyder, etc. ✅ Jupyter Notebooks, Quarto |
Essential Tools for Python Programming
To get the most out of Python–especially for data science and reproducible research–it’s important to set up an integrated, flexible programming environment.
Here is a list of tools we recommend using:
- Conda: a powerful package and environment manager for Python.
- Visual Studio Code: a lightweight code editor that integrates programming + plots + terminal + etc.
- Jupyter Notebook: an interactive computing tool that combines code execution, text documentation, and visualizations.
- Git (GitHub): for version control, collaboration, and publishing code.
Conda (via Miniconda)
Conda is a package manager, much like CRAN + Bioconductor, and can be utilized across languages (Python, R, C/C++ etc.). It also simplifies Python environment management, similar to renv in R but more powerful and flexible, ensuring dependency isolation without cluttering the global system.
Conda can be installed via either Miniconda (lightweight version) or the Anaconda Distribution (full version). We will use the former for the purpose of this workshop series.
Visual Studio Code
Akin to RStudio, Visual Studio Code (often called VS Code) is an IDE for multi-language coding (Python, R, Java, etc.). It has many features integrated within it, including interactive coding via Jupyter Notebook or Quarto, version control with Git, and built-in terminal and debugging tools.
Next, we will walk through installing and setting up these tools.
2. Python Installation and Setup
There are many ways to do install Python locally. For the purpose of the workshop, we recommend one way of installation via Miniconda that works universally across Windows and MacOS platforms.
Miniconda comes with Python, the conda package manager, and a minimal number of libraries. It is a minimal version of the Anaconda Distribution, an open-source distribution of Python designed for scientific computing, data science, machine learning, and AI development. Miniconda is relatively lightweight compared to Anaconda, which adds on top of the Miniconda distribution the Anaconda Navigator graphical user interface (GUI) and over 300 pre-downloaded libraries.
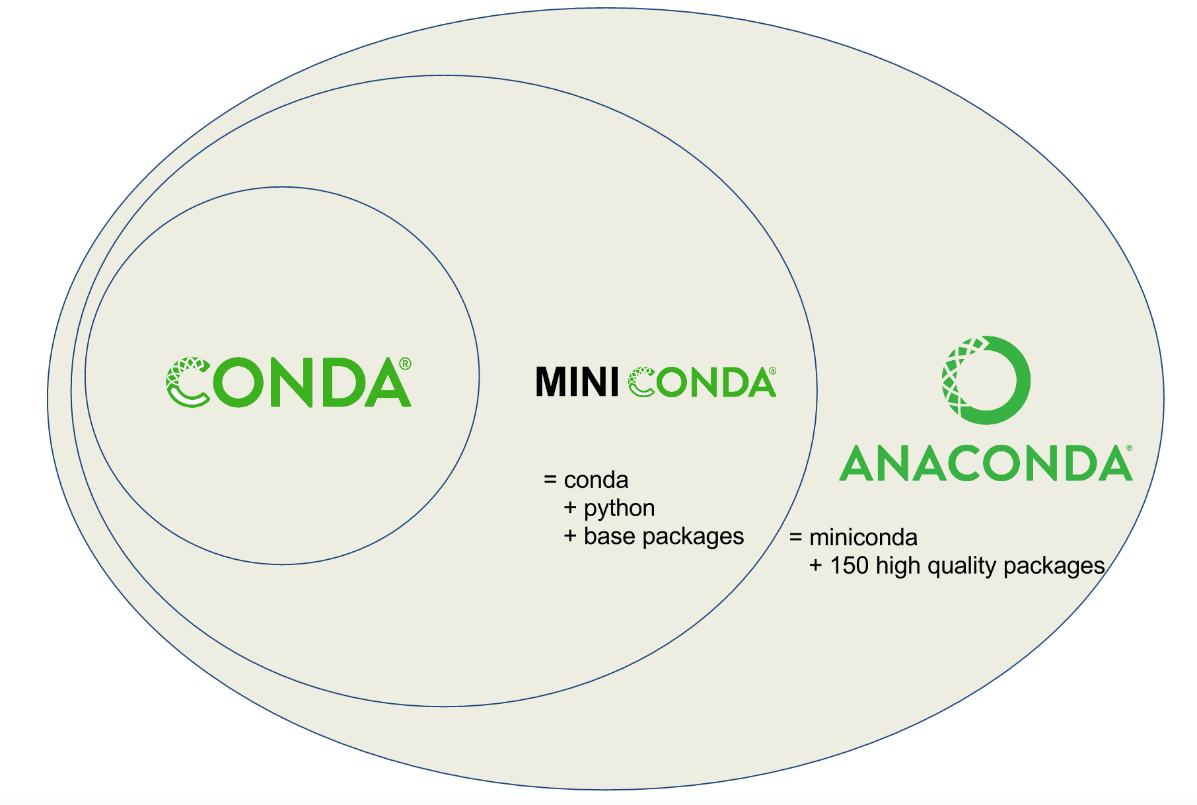
While Anaconda can be an alternative for people who do not want to use command line tools for managing packages, in this workshop, we will use Miniconda for better efficiency in the installation process.
| Feature | Anaconda | Miniconda |
|---|---|---|
| Size | ⚠️~3-4 GB (slow to download) | ✅~400 MB (lightweight) |
| What is included? | conda, Python (latest version), 300+ popular packages, Anaconda Navigator | conda, Python (latest version), essential packages only |
| User-friendly? | ✅GUI available (Anaconda Navigator) | ⚠️command-line only — conda |
▶️Follow-Along: Install Miniconda (Python + conda)
Let’s walk through steps to install Minconda.
For the latest Miniconda installers for Python 3.12, navigate to the Anaconda website.
Download the 64-bit graphical installer according to your system (Windows or MacOS):
Note: Make sure you are downloading from the Miniconda Installers section, not Anaconda!
- Alternatively, check out the Quick command line installation guide to install Miniconda through command line interface.
Run the installer (
.exefor Windows /.pkgfor MacOS)select
Just Mefor installation type – recommended; doesn’t require admin rights.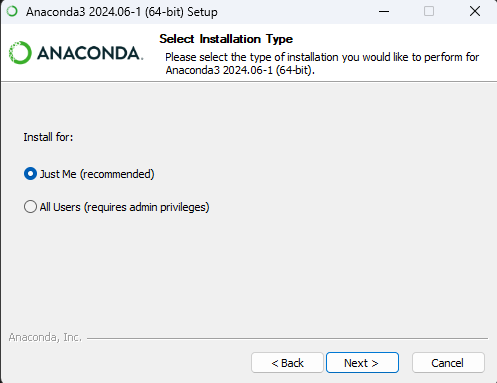 TipInstalling for current user only
TipInstalling for current user onlyYou don’t need to install for all users most of the time. This option requires admin privileges which you might not have on your MSK laptop.
Keep the default for installation location. E.g.,
- Windows:
C:\Users\<user_name>\AppData\Local\miniconda3 - MacOS:
/Users/yourname/miniconda3
- Windows:
Customize the advanced installation options:
- ❌ Add Miniconda to my PATH environment variable – NOT recommended
- ✅ Register Miniconda3 as my default Python 3.12
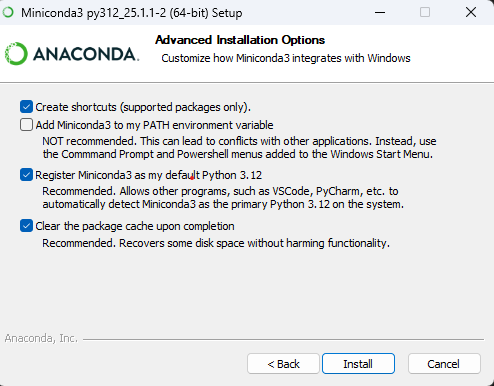
WarningDo not add Miniconda to PATH⚠️It is recommended that you do not add Miniconda to system’s PATH environment variable, as it might lead to conflicts with your other Python installations or accidentally break software using the system Python.
Instead, you could later run
conda initin Anaconda Prompt to configure the terminal shells (like PowerShell or Command Prompt) to recognize thecondacommand.Complete installation. This might take a few minutes to complete.
Check installation–verify that Python and Conda are successfully installed.
Windows:
Open the Start Menu and run Anaconda Prompt.
Type the following command.
conda --version python --versionYou should see the current versions of your Python and Conda being returned–such as
conda 24.9.2andPython 3.12.4(the exact numbers might differ). This means that Miniconda is properly installed and initialized.
MacOS:
Open Terminal. Configure your shell to make the
condacommand available.source ~/miniconda3/bin/activate conda init zsh # or conda init bash if you are using bashThen restart your Terminal and type:
conda --version python --versionYou should see the current versions of your Python and Conda being returned, which means everything has been correctly installed.
3. Virtual Environment & Package Management with Conda
What is Conda?
Conda is a cross-platform command line tool for managing packages and environment via the conda command line interface (CLI). It can handle both Python and non-Python dependencies (R, C, system binaries, etc.), making it particularly powerful.
You can install conda via installers such as Miniconda or the Anaconda Distribution.
Conda for Managing Packages
Conda installs packages from channels(repositories), such as the default Anaconda channel or community-maintained channels like conda-forge.
To install packages from the default Anaconda repository:
# Install a single package
conda install scipy
# Install a specific version of a package
conda install scipy=0.15.0
# Install multiple packages
conda install scipy=0.15.0 pandas matplotlibIf the package is located in another channel, such as conda-forge, you can manually specify the channel when installing the package. For example:
conda install conda-forge::pytorch
# or
conda install pytorch --channel conda-forgeTo update a package:
conda update scipyNote that this automatically updates the package to the highest version supported by the current Python series. For example, Python 3.9 updates to the highest available in the 3.x series.
To remove a package (or multiple packages at once):
conda remove scipy pandas matplotlibConda vs. Pip
If a Python package is not available through any conda channel, consider using the pip package manager:
pip installconda install and pip install
Long story short: Pip installs Python libraries only, while conda can install both Python and non-Python packages (e.g., R, C/C++, system binaries).
It is generally recommended that you only use conda install within a conda environment, as anything installed via pip won’t be recognized by conda and vice versa. Using the two interchangeably might overwrite or break packages and mess up the environment.
What if the Python package is unavailable through conda?
The best practice is to install everything with conda first, then use pip only when the package is not available in conda.
Check out this blog for more information on using pip in a conda environment.
What is a Virtual Environment?
A virtual environment is an isolated, self-contained workspace that includes its own language interpreter and package dependencies. Each environment operates independently, ensuring that projects are isolated from one another and from the system’s global setup.
In the previous section, we installed Python 3.12 via Miniconda and set it as the default (global) Python. However, you might need a different version of Python–say, Python 3.8–or a different set of packages for a particular project. In this case, creating a virtual environment allows you to maintain a completely separate Python setup, including its own Python version and /site-packages folder.
You can create as many environments as needed — ideal for managing multiple projects with different requirements.
Why Use Virtual Environments?
You may find the flexibility of environments beneficial in many cases.
- Avoid Conflicts. Creating virtual environments can help resolve potential conflicts between different projects that might require different Python version or conflicting dependencies. Changes made to one environment won’t affect other projects that use different environments.
- Easy Management. When your work is temporary or that you simply want to experiment things without having to worry about breaking things, you can work within a virtual environment and later delete it when needed.
- Sharing Environment. You can share your Python environment and whole list of dependencies with other people through a copy of the
.ymlfile. - Reproducibility. They work as time capsules, allowing you to come back to an older project at any time later by recreating the virtual environment.
▶️Follow-Along: Create a Conda Virtual Environment
Using conda, we can create, activate, update, export, and remove virtual environments, each with its own Python version and set of packages. Let’s practice creating a virtual environment and installing packages into it using the command line.
The Anaconda Navigator is an alternative to creating conda environments without the terminal skills. However, for users comfortable using the command line tool, we recommend the conda approach for better speed, control, and stability,
Open Anaconda Prompt (Windows) or Terminal (MacOS)
Create the virtual environment. Replace
<env-name>with the name you want to give your environment.conda create --name <env-name>Note: you can use
-n(shorthand) and--nameinterchangeably.To create an environment with a specific Python version:
conda create -n <env-name> python=3.10To create an environment with a specific package(s):
conda create -n <env-name> python=3.10 scipy pandas matplotlibor install packages later with separate commands:
conda create -n <env-name> python=3.10 conda install -n <env-name> scipy pandas matplotlibYou can also install packages from channels other than the
defaults(your can pass multiple channels for the package search):conda install -n <env-name> scipy --channel conda-forge --channel biocondaor explicitly specify the channel from which you want the package to be installed:
conda install -n <env-name> conda-forge::scipyNow, activate your environment.
conda activate <env-name>You can verify that your installation was successful by looking up the list of all current environments on your computer.
conda env listThe default location for the installed conda environments (except for the base conda environment) is
..\miniconda3\envs\<env-name>Deactivate the conda environment.
conda deactivateWarningAvoid activating on top of another virtual environment!Always
conda deactivatefirst before activating another one because environments can be stacked. This can lead to chaos in the packages in both environments.💡Tip: make sure you see
(base)at the beginning of the terminal prompt when you are about to activate an environment.Removing an environment.
Remove by environment name:
conda env remove -n <env-name>Remove by environment folder path:
conda env remove --prefix </path/to/your/env>
👉Create an Environment from an environment.yml File
You can also create a virtual environment from a
.ymlconfiguration file.conda env create -f environment.ymlExample: An environment file contains information about the environment name, channels, and dependencies:
name: python310 channels: - defaults dependencies: - python=3.10 - pandas - numpyNoteDownload the
.ymlfile for this Python workshop series here. This file includes the Python version and required channels and dependencies for completing the workshop sessions.Then activate the new environment:
conda activate python310This way, we can skip the cumbersome steps to set up an environment from scratch and easily recreate an environment shared by others or share our environment settings with others.
| Task | Command |
|---|---|
| Create an environment | conda create --name <env-name> |
| List all environments | conda env list |
| Remove an environment | conda remove --name <env-name> --all |
| List packages in current environment | conda list |
Export environment to .yml |
conda env export > environment.yml |
Recreate environment from .yml file |
conda env create -f environment.yml |
4. Integrated Development Environment
An Integrated Development Environment (IDE) is a software application that brings together everything you need to write and run code and manage projects.
It typically includes:
- A code editor
- Terminal panel
- A compiler or interpreter to execute code
- Debugger and version control integration
For Python, there are many existing IDEs that offer great compatibility and multi-functionality.
| Tool | Description |
|---|---|
| VS Code | Lightweight, powerful IDE (extensible with Python & Jupyter extensions) |
| JupyterLab | Interactive notebooks for analysis & reports |
| Spyder | RStudio-like interface, good for scientific Python |
| PyCharm | Full-featured Python IDE (more for software dev) |
We will use the VS Code IDE for the workshop series.
Visual Studio Code
![]()
Visual Studio Code (VS Code) is one of the most popular open-source code editors with many features.
- Multi-Language Programming. VS Code supports multiple programming languages including Python, R, C/C++, JavaScript, etc.
- Integrated Git Source Control. VS Code automatically recognizes and uses the computer’s Git installation. You can easily track changes, stage, and commit changes to your working branch.
- Variety of Project Development Support. You can add extra features such as language packs, debugging tools, Git/Github features, and remote server connector by installing extensions from the Extension Marketplace.
▶️Follow-Along: Set up Jupyter Notebook in VS Code
We’ll now walk through setting up your Python coding environment in VS Code with full support for virtual environments and Jupyter notebooks.
Step 1: Install Required VS Code Extensions
Launch VS Code.
Open the Extensions panel from the left toolbar (or
Ctrl+Shift+Xon Windows/Cmd+Shift+Xon Mac).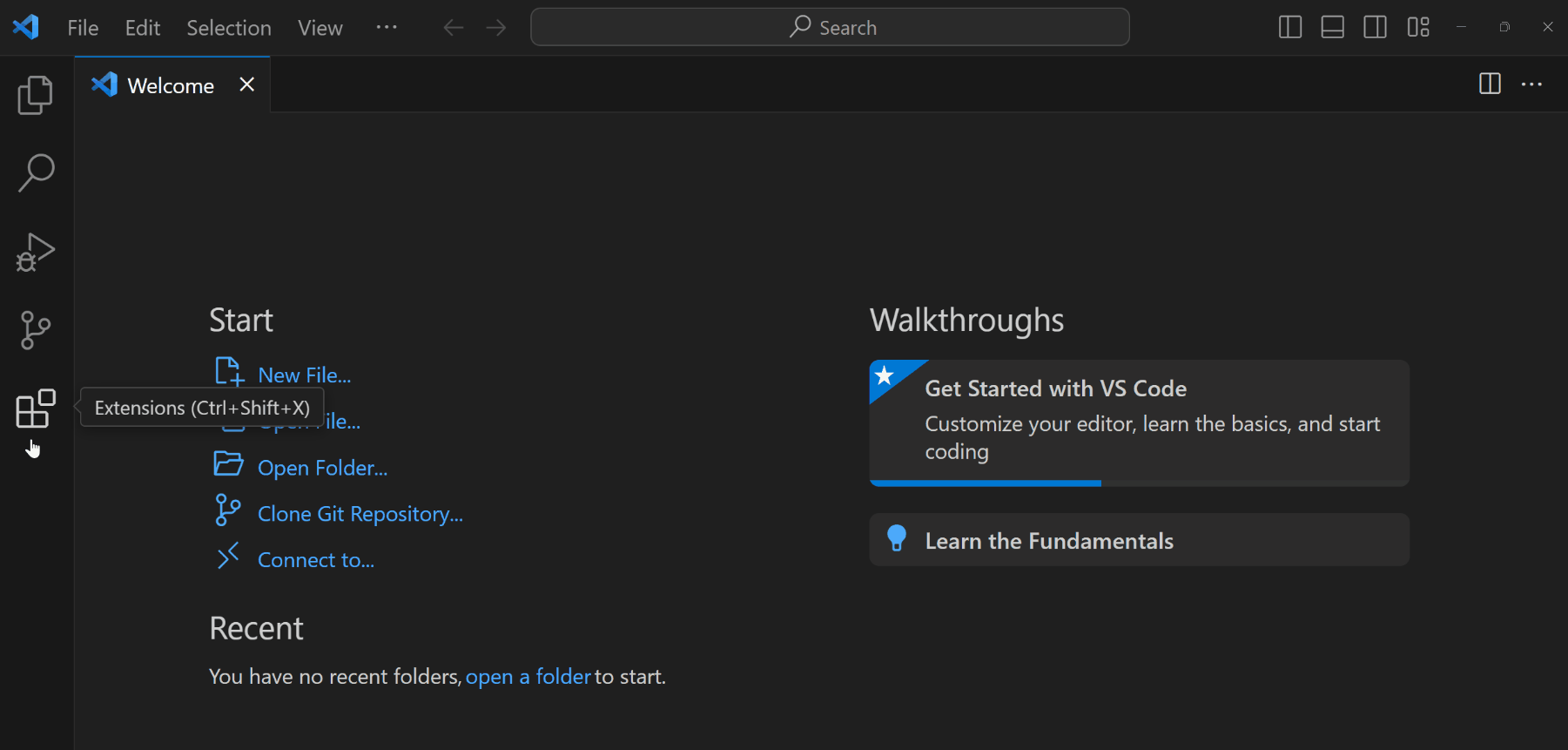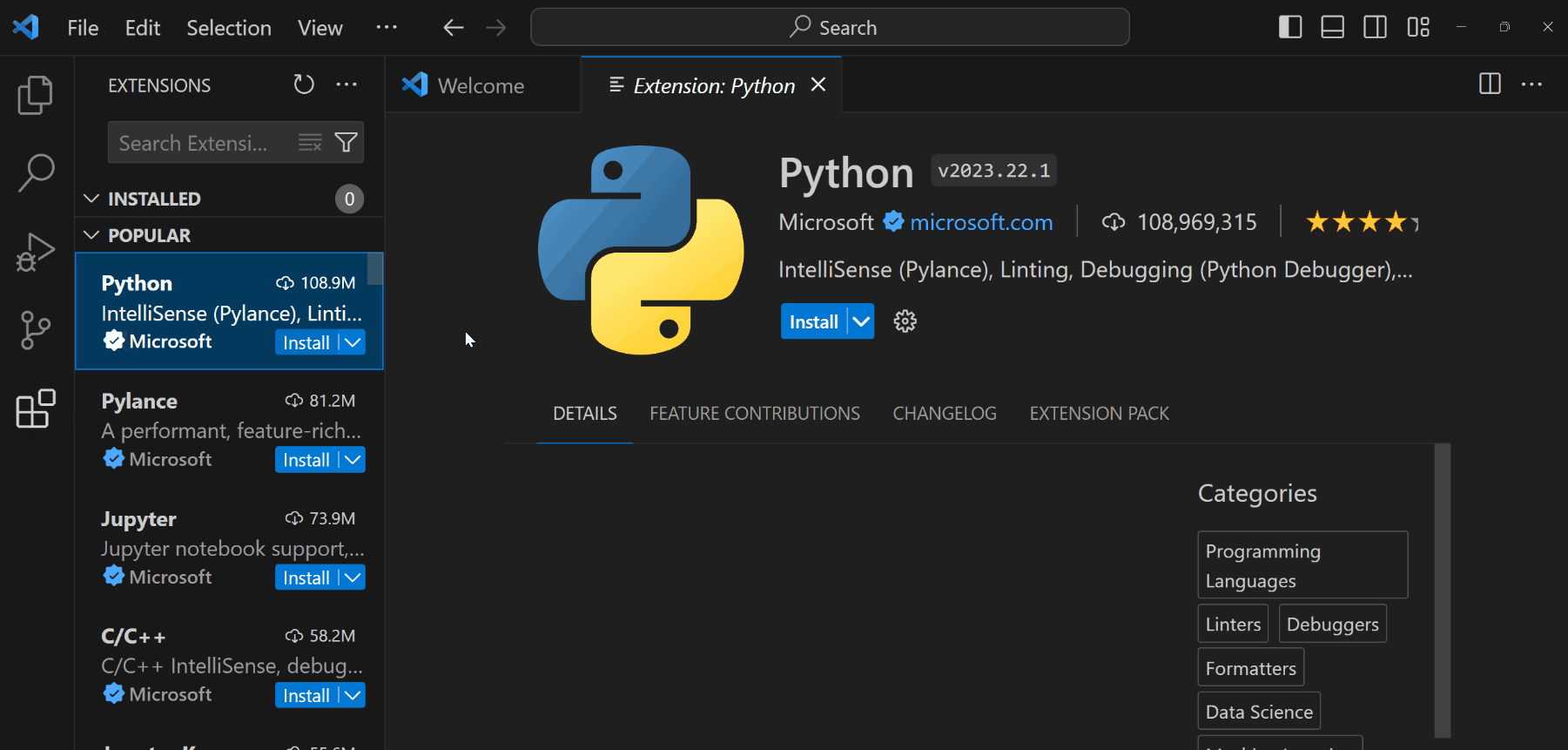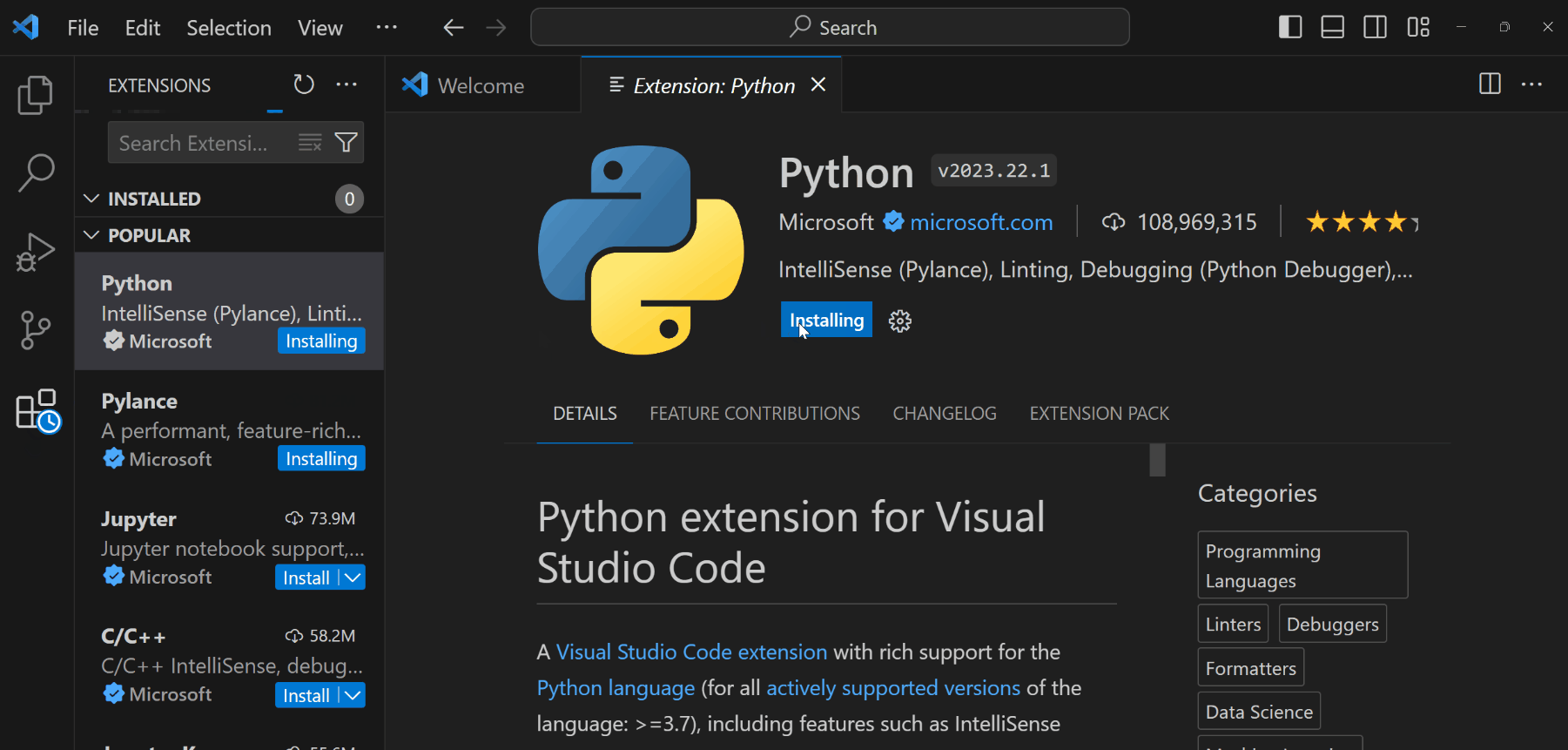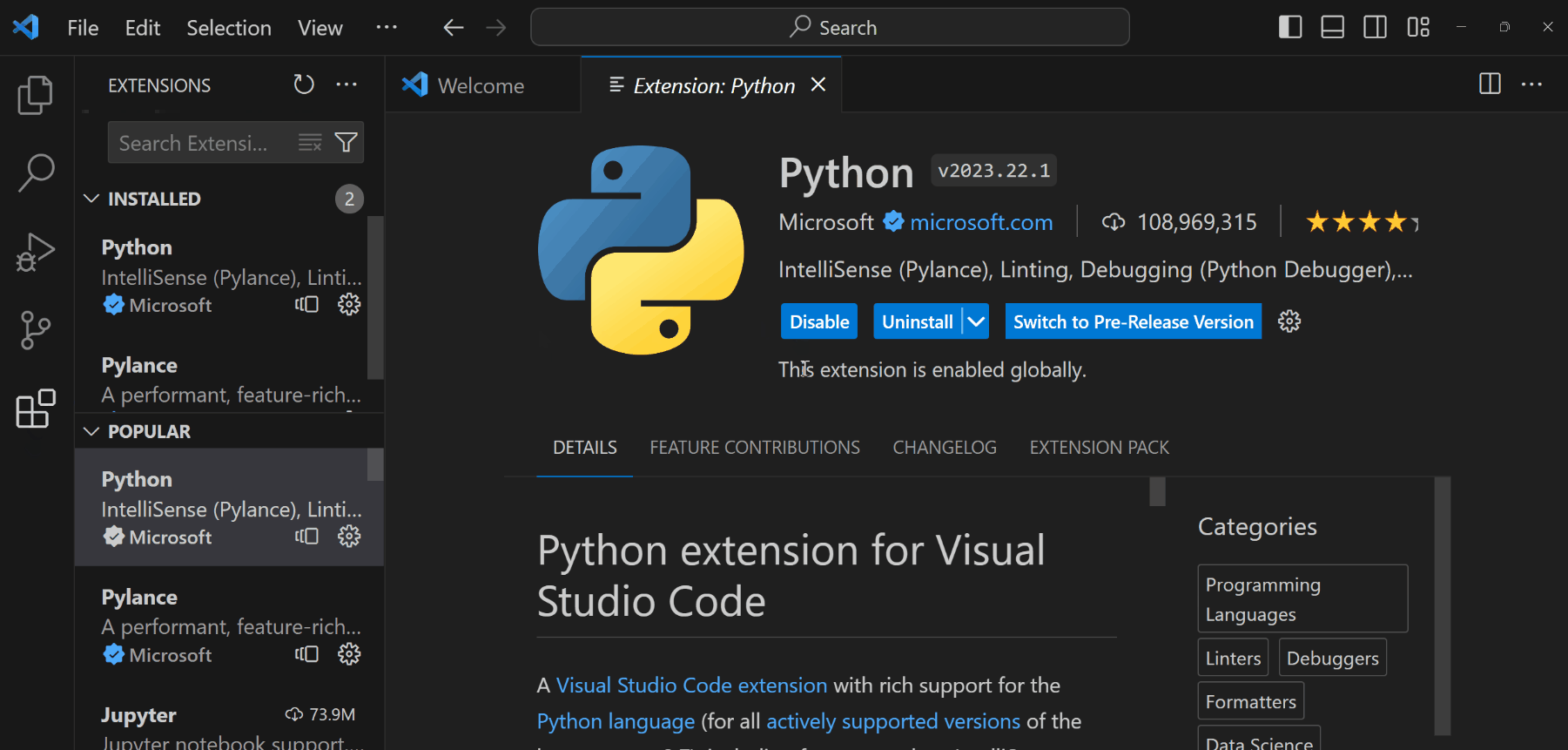
Install the following extensions:
- Python: To support Python language, debugging, documentations, etc.
- Jupyter: To support rendering Python documents from Jupyter Notebooks or Quarto files.
ImportantYou still need to install Python (Miniconda) to your computer for these extensions to fully function.
Step 2: Create a Conda Virtual Environment
You can create an environment in one step using the shared environment.yml file.
conda env create -f environment.ymlIf you want to create your own environment or use existing ones, install these necessary packages to allow your Python to work with Jupyter Notebooks in VS Code:
- jupyter
- ipykernel
- pyyaml
conda install jupyter ipykernel pyyaml -c conda-forgeStep 3: Configure the Environment in VS Code.
Open the Command Palette (
Ctrl+Shift+Pon Windows /Cmd+Shift+Xon Mac).Search for and select “Python: Select Interpreter”.
Choose your conda environment (e.g.,
python-intro-env).If it doesn’t pop up, click Enter interpreter path…
Manually enter the path to your conda virtual environment Python executable:
Windows:
C:\Users\<username>\AppData\Local\miniconda3\envs\<env-name>\python.exemacOS:
Users/<username>/miniconda/envs/<env-name>/python
Step 4: Open a Python Project and Create a Jupyter Notebook File
If you have an existing Python project you wish to work on in VS Code, you may open the project folder in VS Code.
Open from the VS Code Welcome page:

Or by selecting File > Open Folder (
Ctrl+K Ctrl+O)
To create a new Jupyter Notebook file, go to
File > New Fileand select Jupyter Notebook (.ipynb).Select the correct kernel (conda environment):
In the top-right corner of the notebook, you will see a Select Kernel button
Choose the kernel that matches your conda environment (e.g.,
python-intro-env (Python 3.10.0))
If you don’t see the environment showing up:
Make sure your conda env has the dependencies installed:
conda install ipykernel jupyter pyyamlThen restart VS Code or reload the window (
Ctrl+Shift+P>Reload Window).If all packages are installed but the issue persists, manually specify the Python path (see 🔎Find your conda Python executable path)
You are all set!